|
|
|
|
|
|
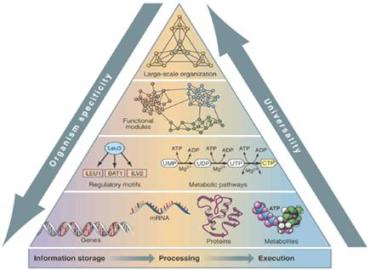
Life's Complexity Pyramid
(Z. N. Oltvai and A.-L. Barabási, Science 298, 763 (2002))


Systems Biology--Metabolic Engineering (BioE 474), Spring semester, 2014
unfinished due to medical leave
Course Description: This Systems Biology course will cover a) the process of reconstructing complex biochemical reaction networks in cells and mathematically simulating their behaviors; b) reconstructing metabolic, gene regulatory and signaling networks; c) bottom-up and top-down approaches; d) principles underlying high-throughput experimental technologies and examples on how this data is used for network reconstruction, consistency checking, and validation; e) classical kinetic theory; and f) constraints-based models. Methods that are scalable and integrate multiple cellular processes will be emphasized. Existing genome-scale models will be described and computations performed. Emphasis will be on studying the genotype-phenotype relationship in an in silico model driven fashion, and comparisons with phenotypic data will be emphasized.
Course Goals: The course will introduce the student to the emerging field of systems biology - the intersection between high-throughput biological data integration and computational modeling. This course will focus primarily on the reconstruction and analysis of biochemical reaction networks (metabolic, regulatory, and signaling) in cells as a basis for systems biology research and synthetic biology designs. By the end of this course, students will be able to:

Course Number
: BioE 474Course Title: Systems Biology- Metabolic Engineering
Cross listed as ChBE 474 - Metabolic Engineering
Credit: 3 hour (undergraduate); 4 hour (graduate)
Semester: Spring 2014
Classroom: 163 Noyes
Lecture Time: 3 hours/week; 2:30-3:50pm WF
Prerequisites: MATH 225, MATH 285, Biology, and Introductory Matlab
Instructor: Charu G. Kumar, Ph.D
E-mail: cgkumar at illinois.edu